Developing a Bioinformatics Database for Disulfide Bonds Research
The Protein Data Bank (PDB) bioinformatics database is the world’s largest repository of experimentally-determined structures of proteins, nucleic acids, and complex assemblies. All data is gathered using experimental methods such as X-ray, spectroscopy, crystallography, NMR, etc. This article explains how to extract, filter, and clean data from the PDB to make it suitable for further analysis.
The Protein Data Bank (PDB) bioinformatics database is the world’s largest repository of experimentally-determined structures of proteins, nucleic acids, and complex assemblies. All data is gathered using experimental methods such as X-ray, spectroscopy, crystallography, NMR, etc. This article explains how to extract, filter, and clean data from the PDB to make it suitable for further analysis.
With a diverse background in science and teaching and his software engineer experience, Victor is a passionate programmer with many talents.
The Protein Data Bank (PDB) bioinformatics database is the world’s largest repository of experimentally-determined structures of proteins, nucleic acids, and complex assemblies. All data is gathered using experimental methods such as X-ray, spectroscopy, crystallography, NMR, etc.
This article explains how to extract, filter, and clean data from the PDB. This, in turn, enables the type of analysis explained in the article Occurrence of protein disulfide bonds in different domains of life: a comparison of proteins from the Protein Data Bank, published in Protein Engineering, Design and Selection, Volume 27, Issue 3, 1 March 2014, pp. 65–72.
The PDB has a lot of repeating structures with different resolutions, methods, mutations, etc. Doing an experiment with the same or similar proteins can produce bias in any group analysis, so we will need to choose the correct structure from among any set of duplicates. For that purpose, we need to use a non-redundant (NR) set of proteins.
For the purpose of normalization, I recommend downloading the chemical compound dictionary for importing atom names into a database that uses 3NF or uses a star schema and dimensional modeling. (I’ve also used DSSP to help eliminate problematic structures. I won’t cover that in this article, but note that I didn’t use any other DSSP features.)
Data used in this research contains single-unit proteins who contain at least one disulfide bond taken from different species. To perform an analysis, all disulfide bonds are first classified as consecutive or nonconsecutive, by domain (archaea, prokaryote, viral, eukaryote, or other), and also by length.
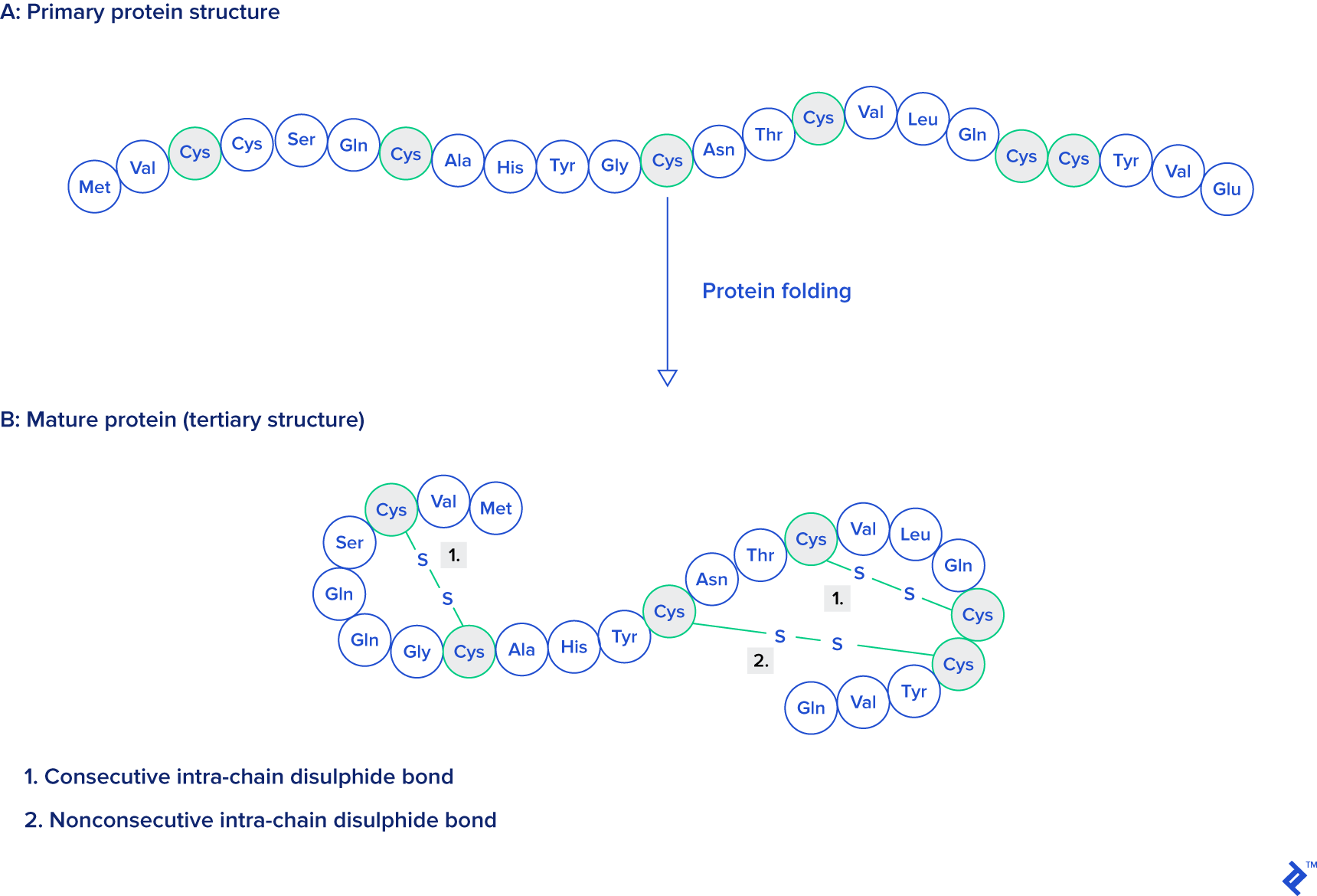
Source: Protein Engineering, Design and Selection, as mentioned at the beginning of this article.
Output
To be ready for input into R, SPSS, or some other tool, an analyst will need the data to be in a database table with this structure:
| Column | Type | Description |
|---|---|---|
code | character(4) | Experiment ID (alphanumeric, case-insensitive, and cannot start with a zero) |
title | character varying(1000) | Title of the experiment, for reference (field can be also text format) |
ss_bonds | integer | Number of disulfide bonds in the chosen chain |
ssbonds_overlap | integer | Number of overlapping disulfide bonds |
intra_count | integer | Number of bonds made within the same chain |
sci_name_src | character varying(5000) | Scientific name of organism from which the sequence is taken |
tax_path | character varying | Path in Linnaean classification tree |
src_class | character varying(20) | Top-level class of organism (eukaryote, prokaryote, virus, archaea, other) |
has_reactives7 | boolean | True if and only if the sequence contains reactive centers |
len_class7 | integer | Length of sequence in set 7 (set with p-value 10e-7 calculated by blast) |
Materials and Methods
In order to accomplish this goal, the first step is to gather data from rcsb.org. That site contains downloadable PDB structures of experiments in various formats.
Although data is stored in multiple formats, in this example, only the formatted fixed-space delimited textual format (PDB) will be used. An alternative to the PDB textual format is its XML version, PDBML, but it sometimes contains malformed atom position naming entries, which can cause problems for data analysis. The older mmCIF and other formats may also be available, but they will not be explained in this article.
The PDB Format
The PDB format is a fragmented fixed-width textual format which can easily be parsed by SQL queries, Java plugins, or Perl modules, for example. Each data type in the file container is represented as a line beginning with the appropriate tag—we’ll go through each tag type in the following subsections. The line length is less than or equal to 80 characters, where a tag takes six or less characters plus one or more spaces which together take eight bytes. There are also cases without spaces between tags and data, usually for CONECT tags.
TITLE
The TITLE tag marks a line as being (part of) the title of the experiment, containing the molecule name and other relevant data like the insertion, mutation, or deletion of a specific amino acid.
12345678901234567890123456789012345678901234567890123456789012345678901234567890
TITLE A TWO DISULFIDE DERIVATIVE OF CHARYBDOTOXIN WITH DISULFIDE
TITLE 2 13-33 REPLACED BY TWO ALPHA-AMINOBUTYRIC ACIDS, NMR, 30
TITLE 3 STRUCTURES
In the case where there are multiple lines to a TITLE record, then the title has to be concatenated, ordering by a continuation number, which is placed, right-aligned, on bytes 9 and 10 (depending on the number of these lines).
ATOM
The data stored in ATOM lines is coordinate data for each atom in an experiment. Sometimes an experiment has insertions, mutations, alternate locations, or multiple models. This results in repeating the same atom multiple times. Choosing the right atoms will be explained later.
12345678901234567890123456789012345678901234567890123456789012345678901234567890
ATOM 390 N GLY A 26 -1.120 -2.842 4.624 1.00 0.00 N
ATOM 391 CA GLY A 26 -0.334 -2.509 3.469 1.00 0.00 C
ATOM 392 C GLY A 26 0.682 -1.548 3.972 1.00 0.00 C
ATOM 393 O GLY A 26 0.420 -0.786 4.898 1.00 0.00 O
ATOM 394 H GLY A 26 -0.832 -2.438 5.489 1.00 0.00 H
ATOM 395 HA2 GLY A 26 0.163 -3.399 3.111 1.00 0.00 H
ATOM 396 HA3 GLY A 26 -0.955 -2.006 2.739 1.00 0.00 H
The example above is taken from the experiment 1BAH. The first column marks the type of record, and the second column is the serial number of the atom. Every atom in a structure has its own serial number.
Next to the serial number there is the atom position label, which takes four bytes. From that atom position, it is possible to extract the chemical symbol of the element, which is not always present in the record data in its own separate column.
After the atom name there is a three-letter residue code. In the case of proteins, that residue corresponds to an amino acid. Next, the chain is coded with one letter. By chain we mean a single chain of amino acids, with or without gaps, although sometimes ligands can be assigned to a chain; this case is detectable through very large gaps in an amino acid sequence, which is in the next column. Sometimes the same structure can be scanned with mutations included, in which case the insertion code is available in an extra column after the sequence column. The insertion code contains a letter to help to distinguish which residue is affected.
The next three columns are the spatial coordinates of each atom, measured in Angstroms (Å). Next to these coordinates is the occupancy column, which says what the probability is for the atom to be in that place, on the usual scale of zero to one.
The second-last column is the temperature factor, which carries information about the disorder in the crystal, measured in Ų. A value greater than 60Ų signifies disorder, while one lower than 30Ų signifies confidence. It is not always present in PDB files because it depends on the experimental method.
The next columns—symbol and charge—are usually missing. The chemical symbol can be gathered from the atom position column, as we mentioned above. When the charge is present, it’s suffixed to the symbol as an integer followed by + or -, e.g. N1+.
TER
This marks the end of the chain. Even without this line, it is easy to distinguish where a chain ends. Thus, often the TER line is missing.
MODEL and ENDMDL
A MODEL line marks where the model of a structure starts, and it contains the serial number of the model.
After all atomic lines in that model, it ends with an ENDMDL line.
SSBOND
These lines contain disulphide bonds between cysteine amino acids. Disulfide bonds can be present in other residue types, but in this article only amino acids will be analyzed, so only cysteine is included. The following example is from the experiment with code 132L:
12345678901234567890123456789012345678901234567890123456789012345678901234567890
SSBOND 1 CYS A 6 CYS A 127 1555 1555 2.01
SSBOND 2 CYS A 30 CYS A 115 1555 1555 2.05
SSBOND 3 CYS A 64 CYS A 80 1555 1555 2.02
SSBOND 4 CYS A 76 CYS A 94 1555 1555 2.02
In this example there are four disulfide bonds tagged in the file with their sequence number in the second column. All of these bonds use cysteine (columns 3 and 6), and all of them are present in chain A (columns 4 and 7). After each chain there is a residue sequence number indicating the bond’s position in the peptide chain. Insertion codes are next to each residue sequence, but in this example they aren’t present because there was no amino acid inserted in that region. The two columns before the last one are reserved for symmetry operations, and the last column is the distance between sulfur atoms, measured in Å.
Let’s take a moment to give some context to this data.
The pictures below, taken using the rcsb.org NGL viewer, show the structure of experiment 132L. In particular, they show a protein without ligands. The first picture uses a stick representation, with CPK coloring showing sulfurs and their bonds in yellow. V-shaped sulfur connections represent methionine connections, while the Z-shaped connections are disulfide bonds between cysteines.
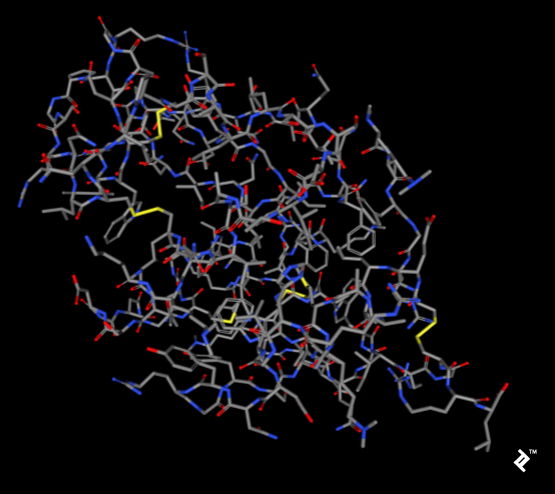
In the next picture, a simplified method of protein visualization called backbone visualization is shown colored by amino acids, where cysteines are yellow. It represents the same protein, with its sidechain excluded, and only part of its peptide group included—in this case, the protein backbone. It’s made of three atoms: N-terminal, C-alpha, and C-terminal. This picture doesn’t show disulfide bonds, but it’s simplified to show the protein’s spatial arrangement:

Pipes are created by joining peptide-bonded atoms with a C-alpha atom. Cysteine’s color is same as the color of sulfur in the CPK coloring method. When cystenes come close enough, their sulfurs create disulfide bonds, and that strengthens the structure. Otherwise this protein would bind too much, and its structure would be less stable at higher temperatures.
CONECT
These records are used for tagging connections between atoms. Sometimes these tags are not present at all, whereas other times all the data is filled in. In the case of analyzing disulfide bonds, this part can be useful—but it isn’t necessary. That’s because, in this project, non-tagged bonds are added by measuring distances, so this would be overhead, and also has to be checked.
SOURCE
These records contain information about the source organism from which the molecule has been extracted. They contain subrecords for easier location in taxonomy, and have the same multi-line structure we saw with title records.
SOURCE MOL_ID: 1;
SOURCE 2 ORGANISM_SCIENTIFIC: ANOPHELES GAMBIAE;
SOURCE 3 ORGANISM_COMMON: AFRICAN MALARIA MOSQUITO;
SOURCE 4 ORGANISM_TAXID: 7165;
SOURCE 5 GENE: GST1-6;
SOURCE 6 EXPRESSION_SYSTEM: ESCHERICHIA COLI;
SOURCE 7 EXPRESSION_SYSTEM_TAXID: 562;
SOURCE 8 EXPRESSION_SYSTEM_STRAIN: BL21(DE3)PLYSS;
SOURCE 9 EXPRESSION_SYSTEM_VECTOR_TYPE: PLASMID;
SOURCE 10 EXPRESSION_SYSTEM_PLASMID: PXAGGST1-6
NR Format
This is a list of non-redundant (NR) chain PDB sets. Its snapshots can be found at ftp.ncbi.nih.gov/mmdb/nrtable/. Its purpose is to avoid unnecessary biases caused by protein similarity. NR has three sets with different identity p-value levels created by comparison of all PDB structures. The result is added to textual files which will be explained later. Not all columns are needed for this project, so only the important ones will be explained.
The first two columns contain the unique PDB experiment code and the chain identifier as explained for ATOM records above. Columns 6, 9, and C contain information about p-value representativity, which is the level of similarity of sequences calculated by BLAST. If that value is zero, then it is not accepted to be part of a set; if the value is 1, then it is. The mentioned columns represent the acceptance of sets with p-values cutoffs of 10e-7, 10e-40, and 10e-80, respectively. Only sets with a p-value cutoff of 10e-7 will be used for analysis.
The last column contains info about a structure’s acceptability, where a is acceptable and n is not.
#---------------------------------------------------------------------------------------------------------------------------
# 1 2 3 4 5 6 7 8 9 A B C D E F G H I J K L M N O P Q
#---------------------------------------------------------------------------------------------------------------------------
3F8V A 69715 1 1 1 1 1 1 1 1 1 9427 1 1 0.00 0.00 0.00 0.00 1.08 1 6 5 164 X a
3DKE X 68132 1 2 0 1 2 0 1 2 0 39139 1 1 0.00 0.00 0.00 0.00 1.25 1 11 7 164 X a
3HH3 A 77317 1 3 0 1 3 0 1 3 0 90 1 0 0.00 0.00 0.00 0.00 1.25 1 5 4 164 X a
3HH5 A 77319 1 4 0 1 4 0 1 4 0 90 2 0 0.00 0.00 0.00 0.00 1.25 1 4 4 164 X a
Database Construction and Parsing Data
Now that we have an idea of what we’re dealing with and what we need to do, let’s get started.
Downloading Data
All data for this analysis can be found at these three addresses:
- ftp.wwpdb.org/pub/pdb/data/structures/divided/pdb/
- ftp.ncbi.nih.gov/mmdb/nrtable/
- ftp.ncbi.nih.gov/pub/taxonomy/taxdmp.zip
The first two links contain a list of archives. The latest archive from each should be used to avoid problems stemming from a lack of resolution or quality. The third link contains the newest taxonomy archive directly.
Parsing Data
Usually the parsing of PDB files is done by plugins or modules in Java, Perl, or Python. In the case of this research, I wrote a custom Perl application without using a pre-written PDB-parsing module. The reason for that is when parsing a large quantity of data, in my experience, the most common problem with using experimental data is errors in the data. Sometimes there are errors with coordinates, distances, line lengths, comments in places where they shouldn’t be, etc.
The most effective way to deal with this is to initially store everything in the database as raw text. Common parsers are written to deal with ideal data that conforms completely to specifications. But in practice, data is not ideal, and that will be explained in filtering section where you’ll find the Perl import script.
Database Construction
When constructing the database, note that this database is built for processing data. Later analysis will be done in SPSS or R. For our purposes here it is recommended to use PostgreSQL with at least version 8.4.
The table structure is directly copied from the downloaded files with only a few small changes. In this case, the number of records is far too small for it to be worth spending our time on normalization. As mentioned, this database is single-use only: These tables aren’t built to be served on a website—they are just there for processing data. Once that is finished, they can be dropped, or backed up as supplementary data, perhaps for repeating the process by some other researcher.
In this case, the final result will be one table which can then be exported to a file for use with some statistical tool like SPSS or R.
Tables
Data extraction from ATOM records has to be connected to HEADER or TITLE records. The data hierarchy is explained in the picture below.
Since this picture is a simplified representation of a database in the third normal form (3NF), for our purposes it contains too much overhead. The reason: To calculate the distance between atoms for disulfide bond detection, we would need to do joins. In this case, we would have a table joined to itself twice, and also joined to a secondary and primary structure twice each, which is a very slow process. Since not every analysis needs secondary structure information, another schema is proposed in case you need to reuse data or analyze bigger quantities of disulfide bonds:
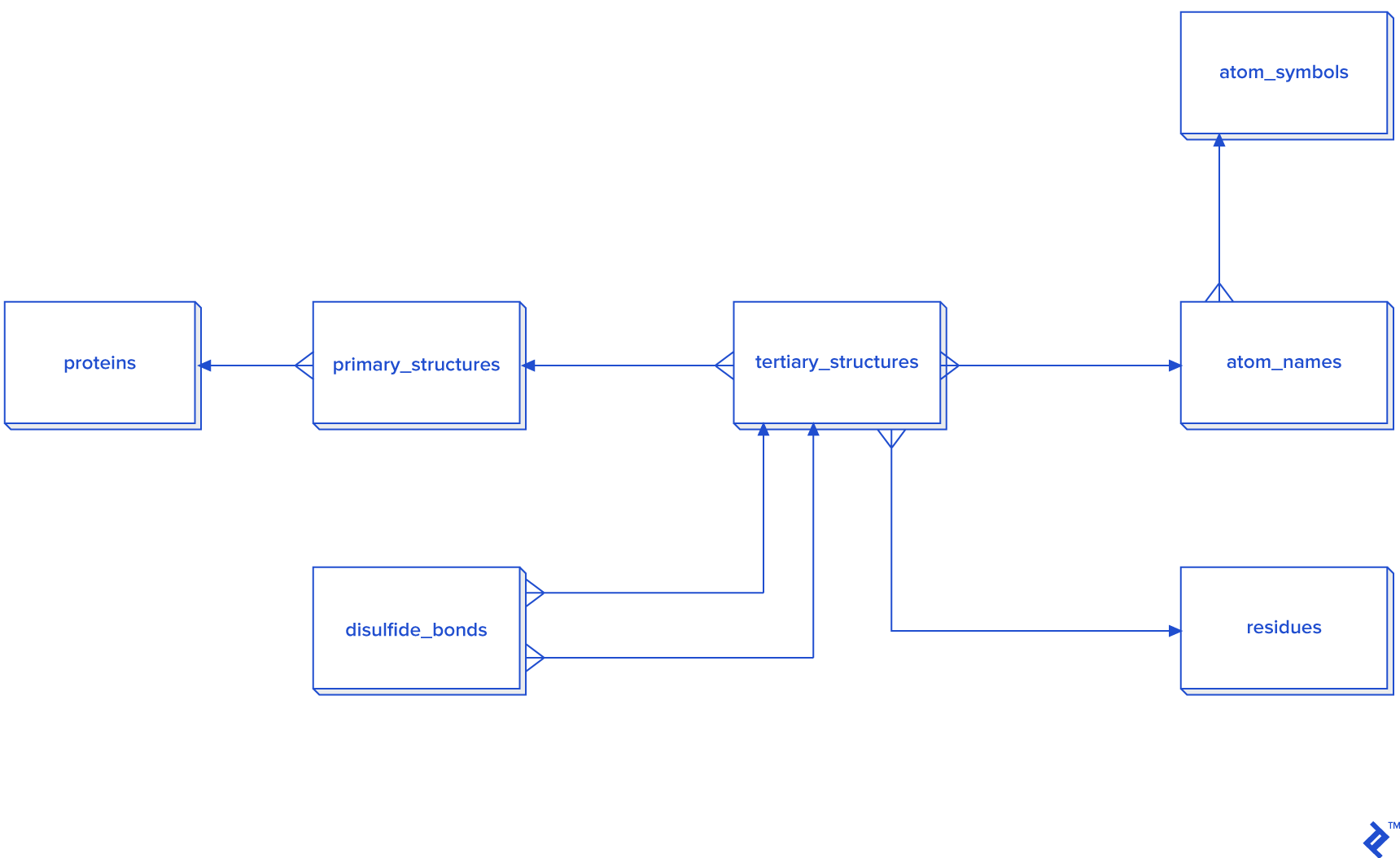
Disulfide bonds are not so frequent as other covalent bonds are, so a warehouse model is not needed, although it could be used. The star schema and dimensional modeling below will take too much time to develop, and will make queries more complex:
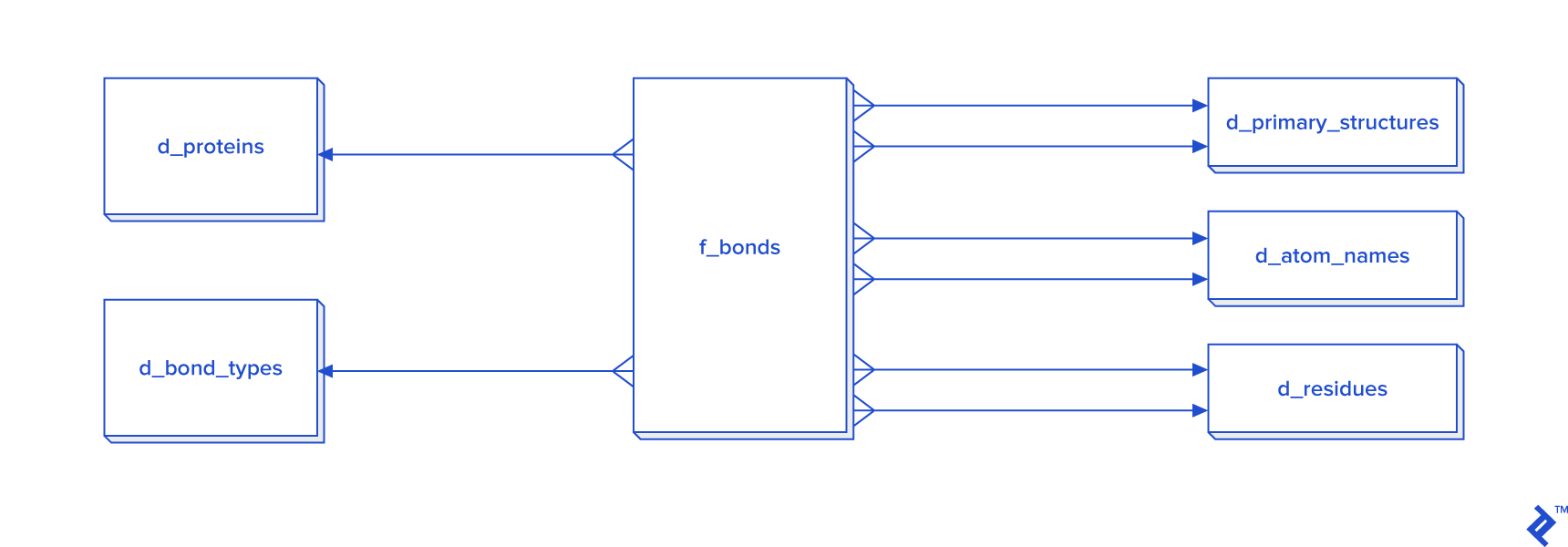
In cases where all bonds have to be processed, then I recommend the star schema.
(Otherwise it’s not needed, because disulfide bonds aren’t as common as other bonds are. In the case of this work, the number of disulfide bonds is near 30,000, which may be fast enough in 3NF, but I decided to process it via a non-normalized table, so it isn’t pictured here.)
The expected total number of all covalent bonds is at least twice the number of atoms in the tertiary structure, and in that case 3NF would be very slow, so denormalization using the star schema form is needed. In that schema, some tables have two foreign key checks, and that is because a bond is created between two atoms, so each atom needs to have its own primary_structure_id, atom_name_id and residue_id.
There are two ways to fill the d_atom_name dimension table: from data, and from another source, the chemical component dictionary I mentioned earlier. Its format is similar to the PDB format: Only RESIDUE and CONECT lines are useful. This is because RESIDUE’s first column contains a residue three-letter code, and CONECT contains the name of the atom and its connections, which are also atom names. So from this file, we can parse all atom names and include them in our database, although I recommend you allow for the possibility of the database containing unlisted atom names.
RESIDUE PRO 17
CONECT N 3 CA CD H
CONECT CA 4 N C CB HA
CONECT C 3 CA O OXT
CONECT O 1 C
CONECT CB 4 CA CG HB2 HB3
CONECT CG 4 CB CD HG2 HG3
CONECT CD 4 N CG HD2 HD3
CONECT OXT 2 C HXT
CONECT H 1 N
CONECT HA 1 CA
CONECT HB2 1 CB
CONECT HB3 1 CB
CONECT HG2 1 CG
CONECT HG3 1 CG
CONECT HD2 1 CD
CONECT HD3 1 CD
CONECT HXT 1 OXT
END
HET PRO 17
HETNAM PRO PROLINE
FORMUL PRO C5 H9 N1 O2
In this project, speed of coding is more relevant than speed of execution and storage consumption. I decided not to normalize—after all, our goal is to generate a table with the columns mentioned in the intro.
In this part, only the most important tables will be explained.
The main tables are:
-
proteins: Table with experiment names and codes. -
ps: Primary structure table which will containsequence,chain_id, andcode. -
ts: Table containing tertiary/quaternary structure extracted from raw data and transformed intoATOMrecord format. This will be used as a staging table, and can be dropped after extraction. Ligands are excluded. -
sources: The list of organisms from which experimental data was derived. -
tax_names,taxonomy_path,taxonomy_paths: Linnean taxonomy names from the NCBI taxonomy database, used to get taxonomy paths from organisms listed insources. -
nr: List of NCBI non-redundant proteins extracted from the NR set. -
pdb_ssbond: List of disulfide bonds in a given PDB file.
Filtering and Processing Data
Data is retrieved from snapshots from the RCSB PDB repository.
Each file is imported into a single table raw_pdb in our PostgreSQL database using a Perl script. The script uses transactions of 10,000 inserts per chunk.
The structure of raw_pdb is this:
| Column | Type | Modifiers |
|---|---|---|
| code | character varying(20) | not null |
| line_num | integer | not null |
| line_cont | character varying(80) | not null |
The import script looks like this:
#!/usr/bin/perl -w
use FindBin '$Bin';
use DBI;
$dbName = 'bioinf';
$dbLogin = 'ezop';
$dbPass = 'XYZ';
$conn = DBI->connect("DBI:Pg:database=$dbName;host=localhost", "$dbLogin", "$dbPass", {'RaiseError' => 1, 'AutoCommit' => 0});
die "./pdb_lines_unos.pl <table> <file>" if not defined($ARGV[0]);
$recordCount = 0;
$table = $ARGV[0];
$fName = $ARGV[1];
open F, "zcat $fName|";
while (<F>) {
chomp;
$linija = $_;
$recordCount += 1;
$sql = "insert into $table (code, line_num, line_cont) values (?, ?, ?)";
$conn->do($sql, undef, $fName, $recordCount, $linija);
$conn->commit() if ($recordCount%10000 == 0);
}
close F;
$conn->commit();
1;
After lines are imported, they are parsed using functions we will define below.
From raw_pdb data, we generate the tables ts, ps, proteins, sources, sources_organela, and ss_bond by parsing the corresponding records.
The ps table has three important columns: chain, length, and sequence. Protein sequence is generated using C-alpha atoms for each chain and for each residue ordered by residue sequence, taking only the first insertion and the first alternate location. chain is taken from the TS.chain column, and length is simply the precalculated length of the sequence string. Since this article is meant to analyze only single chains and intrachain connections, multiple-chain proteins are skipped in our analysis here.
Within SSBOND records, all disulfide bonds are stored in the pdb_ssbond table, which inherits from the pdb_ssbond_extended table. pdb_ssbond looks like this:
| Column | Type | Nullable | Default | Description |
|---|---|---|---|---|
| id | integer | not null | nextval(‘pdb_ssbond_id_seq’::regclass) | |
| code | character(4) | four-letter code | ||
| ser_num | integer | serial number of ssbond | ||
| residue1 | character(3) | first residue in bond | ||
| chain_id1 | character(1) | first chain in bond | ||
| res_seq1 | integer | sequential number of first residue | ||
| i_code1 | character(1) | insertion code of first residue | ||
| residue2 | character(3) | second residue in bond | ||
| chain_id2 | character(1) | second chain in bond | ||
| res_seq2 | integer | sequential number of second residue | ||
| i_code2 | character(1) | insertion code of second residue | ||
| sym1 | character(6) | first symmetry operator | ||
| sym2 | character(6) | second symmetry operator | ||
| dist | numeric(5,2) | distance between atoms | ||
| is_reactive | boolean | not null | false | mark for reactivity (to be set) |
| is_consecutive | boolean | not null | true | mark for consecutive bonds (to be set) |
| rep7 | boolean | not null | false | mark for set-7 structures (to be set) |
| rep40 | boolean | not null | false | mark for set-40 structures (to be set) |
| rep80 | boolean | not null | false | mark for set-80 structures (to be set) |
| is_from_pdb | boolean | true | is taken from PDB as source (to be set) |
I also added these indexes:
"pdb_ssbond_pkey" PRIMARY KEY, btree (id)
"ndxcode1" btree (code, chain_id1, res_seq1)
"ndxcode2" btree (code, chain_id2, res_seq2)
It is assumed that distribution of disulfide bonds prior to the cutoff is Gaussian (without testing with, e.g., KS-test), so standard deviations were calculated on each distance between cysteines in same protein to discover the range of permitted bond lengths and compare them to the cutoff. The cutoff was same as the calculated mean plus-minus three standard deviations. We have extended the range to introduce more possible disulfide bonds which weren’t enlisted in PDB file in SSBOND rows but which we have inserted ourselves by calculating distances between ATOM records. The chosen range for ssbonds are between 1.6175344456264 and 2.48801947642267 Å, which is the mean (2.05) plus-minus four standard deviations:
select count(1) as cnt
, stddev(dist) as std_dev
, avg(dist) as avg_val
, -stddev(dist) + avg(dist) as left1
, stddev(dist) + avg(dist) as right1
, -2 * stddev(dist) + avg(dist) as left2
, 2 * stddev(dist) + avg(dist) as right2
, -3 * stddev(dist) + avg(dist) as left3
, 3 * stddev(dist) + avg(dist) as right3
, -4 * stddev(dist) + avg(dist) as left4
, 4 * stddev(dist) + avg(dist) as right4
, min(dist)
, max(dist)
from pdb_ssbond_tmp
where dist > 0
and dist < 20;
The TS table contains the coordinates of all atoms, but only cysteines will be used, with their sulfur named " SG ". So another staging table with " SG " sulfur atoms only is created for speeding up the process by reducing the number of records to search. When selecting sulfurs only, the number of combinations is much less than in the case of all atoms—194,574 of the former compared with 122,761,100 of the latter. Within this table joined to itself, distances are calculated using the Euclidean distance, and results are imported into the pdb_ssbond table but only where the distance is between the defined lengths calculated earlier. The reason for doing this speedup is to lessen the amount of time of running the whole process again—for checking purposes—keeping it within one day.
To clean disulfide bonds, we use the following algorithm:
- Delete when both sides of connection point to same amino acid
- Delete bonds whose length is not between 1.6175344456264 and 2.48801947642267
- Remove insertions
- Remove bonds caused by alternate atom locations, but leaving first location
The code for this (taking the pdb_ssbond table as the first argument) is:
CREATE OR REPLACE FUNCTION pdb_ssbond_clean2(
clean_icodes boolean,
clean_altloc boolean,
mark_reactive boolean,
mark_consecutive boolean)
RETURNS void AS $$
declare _res integer;
BEGIN
delete from pdb_ssbond b
where exists (
select a.id
from pdb_ssbond a
where a.code = b.code
and a.id > b.id
and (
( a.chain_id1 = b.chain_id1 and a.res_seq1 = b.res_seq1
and a.chain_id2 = b.chain_id2 and a.res_seq2 = b.res_seq2)
or
( a.chain_id1 = b.chain_id2 and a.res_seq1 = b.res_seq2
and a.chain_id2 = b.chain_id1 and a.res_seq2 = b.res_seq1)
)
) ;
delete
from pdb_ssbond
where chain_id1 = chain_id2
and res_seq1 = res_seq2
and i_code1 = i_code2;
update pdb_ssbond
set is_consecutive = true
, is_reactive = false;
delete from pdb_ssbond
where not dist between 1.6175344456264 and 2.48801947642267;
if clean_icodes then
delete from pdb_ssbond
where i_code1 not in ('', ' ', 'A')
or i_code2 not in ('', ' ', 'A') ;
end if;
if clean_altloc then
delete from pdb_ssbond a
where exists (
select 1
from pdb_ssbond b
where b.code = a.code
and b.chain_id1 = a.chain_id1
and b.res_seq1 = a.res_seq1
and b.i_code1 = a.i_code1
and b.ser_num < a.ser_num
) ;
delete from pdb_ssbond a
where exists (
select 1
from pdb_ssbond b
where b.code = a.code
and b.chain_id2 = a.chain_id2
and b.res_seq2 = a.res_seq2
and b.i_code2 = a.i_code2
and b.ser_num < a.ser_num
) ;
end if;
if mark_reactive then
update pdb_ssbond
set is_reactive = false ;
update pdb_ssbond
set is_reactive = true
where exists (
select 1
from pdb_ssbond b
where b.code = pdb_ssbond.code
and b.chain_id1 = pdb_ssbond.chain_id1
and b.res_seq1 = pdb_ssbond.res_seq1
and b.i_code1 = pdb_ssbond.i_code1
and b.ser_num < pdb_ssbond.ser_num
) ;
update pdb_ssbond
set is_reactive = true
where exists (
select 1
from pdb_ssbond b
where b.code = pdb_ssbond.code
and b.chain_id2 = pdb_ssbond.chain_id2
and b.res_seq2 = pdb_ssbond.res_seq2
and b.i_code2 = pdb_ssbond.i_code2
and b.ser_num < pdb_ssbond.ser_num
) ;
update pdb_ssbond
set is_reactive = true
where (code, chain_id1, res_seq1, i_code1)
in (
select code, chain_id1 as c, res_seq1 as r, i_code1 as i
from pdb_ssbond
intersect
select code, chain_id2 as c, res_seq2 as r, i_code2 as i
from pdb_ssbond
) ;
update pdb_ssbond
set is_reactive = true
where (code, chain_id2, res_seq2, i_code2)
in (
select code, chain_id1 as c, res_seq1 as r, i_code1 as i
from pdb_ssbond
intersect
select code, chain_id2 as c, res_seq2 as r, i_code2 as i
from pdb_ssbond
);
end if;
if mark_consecutive then
update pdb_ssbond
set is_consecutive = false;
update pdb_ssbond
set is_consecutive = true
where not exists (
select 1
from pdb_ssbond a
where
a.code = pdb_ssbond.code
and (
(a.chain_id1 = pdb_ssbond.chain_id1 and a.chain_id2 = pdb_ssbond.chain_id2)
or
(a.chain_id1 = pdb_ssbond.chain_id2 and a.chain_id2 = pdb_ssbond.chain_id1)
)
and (
(a.res_seq1 between pdb_ssbond.res_seq1 and pdb_ssbond.res_seq2)
or (a.res_seq2 between pdb_ssbond.res_seq1 and pdb_ssbond.res_seq2)
or (pdb_ssbond.res_seq1 between a.res_seq1 and a.res_seq2)
or (pdb_ssbond.res_seq2 between a.res_seq1 and a.res_seq2)
)
and not (
a.code = pdb_ssbond.code
and a.chain_id1 = pdb_ssbond.chain_id1
and a.chain_id2 = pdb_ssbond.chain_id2
and a.res_seq1 = pdb_ssbond.res_seq1
and a.res_seq2 = pdb_ssbond.res_seq2
)
);
end if;
END;
$$ LANGUAGE plpgsql;
With this, the non-redundant set of proteins is imported to the nr table which is joined to the ps and proteins tables, and sets are marked (set7, set40, and set80). At the end, according to protein quantity only one set will be analyzed. Mismatched chains between PDB and NR are removed from analysis.
Proteins without disulfide bonds are excluded from research, together with proteins that don’t belong to any set. Data is processed with DSSP, and these files which had problems with resolution or too many atoms to be processed are also excluded. Only proteins with single chains are used as result for analysis because interchain connections were not kept, although they are easily calculated from the ssbond table by counting the number of connections that have different chains.
For the remaining proteins, an update is done for the total number of bonds and the number of overlapping bonds, and this is done for each of the sets.
The source organism is extracted from SOURCE records. In cases where it is unknown, synthetic, designed, artificial, or hybrid, it is discarded from research. Low-resolution proteins are also excluded only when their side chain is not visible.
SOURCE records are stored in the sources table, which contains taxonomy rows. In some cases, the taxonomy is missing or incorrect. In these cases, the manual correction of experts is needed.
From the source and taxonomy downloaded from NCBI, the class is assigned to each primary structure. In case a class is unable to be assigned, the protein is removed from the analysis list. Knowing that biological databases are being used, an extra check of all source records and NCBI taxonomy classes is recommended to be performed by a biologist, otherwise there might be problems with classifications between different domains. To match source cellular locations with taxonomy IDs, data from the source table is extracted into the table sources_organela where all data is written as code, tag, and value. Its format is shown below:
select * from sources_organela where code = '1rav';
| code | mol_id | tag | val |
|---|---|---|---|
| 1rav | 0 | MOL_ID | 1 |
| 1rav | 7 | CELLULAR_LOCATION | CYTOPLASM (WHITE) |
The taxonomy archive we want to use is a ZIP file containing seven dump files. Among these files, two of the most important are names.dmp and merged.dmp. Both files are CSV tab-pipe delimited files as detailed in the documentation:
- The file
merged.dmpcontains a list of previous taxonomy IDs, and the current taxonomy IDs into which each one was merged. -
names.dmpcontains these important columns in this order:-
tax_id: The taxonomy ID -
name_txt: The name of the species, and if applicable, the unique name (where species can be found with multiple names, this helps disambiguate)
-
-
division.dmpcontains the names of top-level domains which we will use as our classes. -
nodes.dmpis the table which contains a hierarchical structure of organisms using taxonomy IDs.- It contains a parent taxonomy ID, a foreign key which can be found in
names.dmp. - It also contains a division ID which is important for us to correctly store the relevant top domain data.
- It contains a parent taxonomy ID, a foreign key which can be found in
With all this data and manual corrections (setting correct domains of life) we were able to create the structure of the taxonomy_path table. A sampling of data looks like this:
select * from taxonomy_path order by length(path) limit 20 offset 2000;
| tax_id | path | is_viral | is_eukaryote | is_archaea | is_other | is_prokaryote |
|---|---|---|---|---|---|---|
| 142182 | cellular organisms;Bacteria;Gemmatimonadetes | f | f | f | f | t |
| 136087 | cellular organisms;Eukaryota;Malawimonadidae | f | t | f | f | f |
| 649454 | Viruses;unclassified phages;Cyanophage G1168 | t | f | f | f | f |
| 321302 | Viruses;unclassified viruses;Tellina virus 1 | t | f | f | f | f |
| 649453 | Viruses;unclassified phages;Cyanophage G1158 | t | f | f | f | f |
| 536461 | Viruses;unclassified phages;Cyanophage S-SM1 | t | f | f | f | f |
| 536462 | Viruses;unclassified phages;Cyanophage S-SM2 | t | f | f | f | f |
| 77041 | Viruses;unclassified viruses;Stealth virus 4 | t | f | f | f | f |
| 77042 | Viruses;unclassified viruses;Stealth virus 5 | t | f | f | f | f |
| 641835 | Viruses;unclassified phages;Vibrio phage 512 | t | f | f | f | f |
| 1074427 | Viruses;unclassified viruses;Mouse Rosavirus | t | f | f | f | f |
| 1074428 | Viruses;unclassified viruses;Mouse Mosavirus | t | f | f | f | f |
| 480920 | other sequences;plasmids;IncP-1 plasmid 6-S1 | f | f | f | t | f |
| 2441 | other sequences;plasmids;Plasmid ColBM-Cl139 | f | f | f | t | f |
| 168317 | other sequences;plasmids;IncQ plasmid pIE723 | f | f | f | t | f |
| 536472 | Viruses;unclassified phages;Cyanophage Syn10 | t | f | f | f | f |
| 536474 | Viruses;unclassified phages;Cyanophage Syn30 | t | f | f | f | f |
| 2407 | other sequences;transposons;Transposon Tn501 | f | f | f | t | f |
| 227307 | Viruses;ssDNA viruses;Circoviridae;Gyrovirus | t | f | f | f | f |
| 687247 | Viruses;unclassified phages;Cyanophage ZQS-7 | t | f | f | f | f |
Before any analysis, to avoid biases, sequences have to be checked for their level of identity. Although the NR set contains sequences which are already compared between each other, an extra check is always recommended.
For each disulfide bond’s prior statistical analysis, data is marked if it is reactive or overlapping. After marking overlaps, that information automatically reveals how many consecutive and non-consecutive bonds are inside each protein, and that data is stored in the proteins table from which all protein complexes are excluded in final result.
Each disulfide bond is marked also for its association to sets, by checking both bond sides to see if they belong to the same NR set. Where that is not the case, the disulfide bond is skipped for that set analysis.
To analyze the quantity of bonds by their variance, each length has to be put in a specific class. In this case, only five classes are chosen as written in the function below.
CREATE OR REPLACE FUNCTION len2class(len integer)
RETURNS integer AS
$BODY$
BEGIN
return
case
when len <= 100 then 1
when len > 100 and len <= 200 then 2
when len > 200 and len <= 300 then 3
when len > 300 and len <= 400 then 4
else 5
end;
END;
$BODY$
LANGUAGE plpgsql VOLATILE
COST 100;
Most of the protein sizes are less than 400 amino acids, so length classification is done by splitting lengths into five ranges, each taking 100 amino acids, except the last one which takes the rest. Below you can see how to use this function to extract data for analysis:
SELECT p.code,
p.title,
p.ss_bonds,
p.ssbonds_overlap,
p.intra_count,
p.sci_name_src,
p.len,
p.tax_path,
p.pfam_families,
p.src_class,
( SELECT s.id
FROM src_classes s
WHERE s.src_class::text = p.src_class::text) AS src_class_id,
p.len_class7,
( SELECT s.val
FROM sources_organela s
WHERE s.code = p.code::bpchar AND s.tag::text = 'EXPRESSION_SYSTEM_CELLULAR_LOCATION'::text) AS expression_system_cellular_location,
( SELECT s.val
FROM sources_organela s
WHERE s.code = p.code::bpchar AND s.tag::text = 'CELLULAR_LOCATION'::text) AS cellular_location,
ps.sequence,
ps.uniprot_code,
ps.accession_code,
ps.cc,
ps.seq_uniprot,
ps.chain_id
FROM proteins p
JOIN nr n ON n.code::text = p.code::text AND n.rep7 = true
JOIN ps ps ON ps.code::text = n.code::text AND ps.chain_id = n.chain_id AND ps.het = false
WHERE p.is_excluded = false AND p.chain_cnt = 1 AND p.is_set7 = true AND p.reactive_cnt7 = 0
ORDER BY p.code;
An example result with corrected titles and some manually added columns is shown below.
| PDB code | Total number of SS-bonds | Number of non-consecutive SS-bonds | PDB length / amino acids | Domain | TargetP 1.1 | TatP 1.0 | SignalP 4.1 | ChloroP 1.1 | TMHMM 2.0 number of transmembrane domains | Big-pi | nucPred | NetNES 1.1 | PSORTb v3.0 | SecretomeP 2.0 | LocTree2 | Consensus localization prediction |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1akp | 2 | 0 | 114 | Bacteria | ND | Tat-signal | no signal peptide | ND | 0 | ND | ND | ND | unknown | ND | fimbrium | unknown |
| 1bhu | 2 | 0 | 102 | Bacteria | ND | Tat-signal | signal peptide | ND | 1 | ND | ND | ND | unknown | ND | secreted | unknown |
| 1c75 | 0 | 0 | 71 | Bacteria | ND | Tat-signal | no signal peptide | ND | 0 | ND | ND | ND | cytoplasmic membrane | nonclassical secretion | periplasm | unknown |
| 1c8x | 0 | 0 | 265 | Bacteria | ND | Tat-signal | signal peptide | ND | 1 | ND | ND | ND | unknown | ND | secreted | unknown |
| 1cx1 | 1 | 0 | 153 | Bacteria | ND | Tat-signal | signal peptide | ND | 1 | ND | ND | ND | extracellular | ND | secreted | unknown |
| 1dab | 0 | 0 | 539 | Bacteria | ND | Tat-signal | signal peptide | ND | 0 | ND | ND | ND | outer membrane | ND | outer membrane | unknown |
| 1dfu | 0 | 0 | 94 | Bacteria | ND | Tat-signal | no signal peptide | ND | 0 | ND | ND | ND | cytoplasmic | ND | cytosol | unknown |
| 1e8r | 2 | 2 | 50 | Bacteria | ND | Tat-signal | signal peptide | ND | 0 | ND | ND | ND | unknown | ND | secreted | secreted |
| 1esc | 3 | 0 | 302 | Bacteria | ND | Tat-signal | signal peptide | ND | 1 | ND | ND | ND | extracellular | ND | periplasm | unknown |
| 1g6e | 1 | 0 | 87 | Bacteria | ND | Tat-signal | signal peptide | ND | 1 | ND | ND | ND | unknown | ND | secreted | unknown |
PostgreSQL as a Processing Intermediary
In this work, we showed how to process data, from fetching to analyzing. When working with scientific data, sometimes normalization is needed, and sometimes not. When working with small quantities of data which will not be reused for another analysis, then it is enough to leave it denormalized where processing is fast enough.
The reason why this was done all in one bioinformatics database is that PostgreSQL is able to integrate many languages. This includes R, which can do statistical analysis directly in-database—the subject for a future article on bioinformatics tools.
A special thanks to Toptal colleagues Stefan Fuchs and Aldo Zelen for their invaluable consultations.
Further Reading on the Toptal Blog:
Understanding the basics
What is a database?
A database is data that’s electronically organized and stored for easier retrieval.
What are the two main types of databases?
In terms of bioinformatics, amino acid databases and nucleic acid databases are the two main types, but there are also hybrids.
What is a disulfide bond in biology?
A disulfide bond is a bond between two sulfur atoms (in this case, between cysteines). Additionally, disulfide bonds can be formed between two cysteines from different protein chains.
What is the role of a disulfide bond?
In the case of proteins, a disulfide bond stabilizes the structure.
Is a disulfide bond polar covalent?
A disulfide bond is a covalent bond. When both sides of the bond are symmetric (both residues are cysteines) then it is non-polar. In the case where some metal ion is near it, the electron cloud changes and it becomes polar like it would be if it were asymmetric. Polar means it attracts water.
Which protein structure has disulfide bonds?
Beta sheets, beta barrels, and beta strands have disulfide bonds. It is possible to find it in other structures, too.
What is the purpose of a database?
To store and organize data for easier retrieval. In this article, a database helps perform ETL (extraction, transformation, and loading) with a relatively large quantity of data. Data is stored in the database, processed inside the database, and then extracted to be analyzed with SPSS or R.
How can bioinformatics be used?
It’s mostly in medicine and biology, but also in physics and chemistry. It can help better understand the structures and properties of different pharmaceuticals, diseases, and abnormalities. It’s used to explore organism functioning, help to recognize unknown species, and find the closest species to a given one.
What is meant by data cleaning?
Biologists’ databases are constantly curated, but often human and machine errors still happen, e.g., impossibly long disulfide bonds, wrongly classified organisms, or wrong taxonomy numbers. The bigger the database, the more errors can be found. One technique used as a starting point is searching for outliers.
About the author
With a diverse background in science and teaching and his software engineer experience, Victor is a passionate programmer with many talents.

